Article Details
QSAR Research of Novel Tetrandrine Derivatives against Human Hepatocellular Carcinoma
Author(s):
Meng Wang, Bin Qiu, Wenhui Wang, Xiang Li and Huixia Huo* Pages 2146 - 2153 ( 8 )
Abstract:
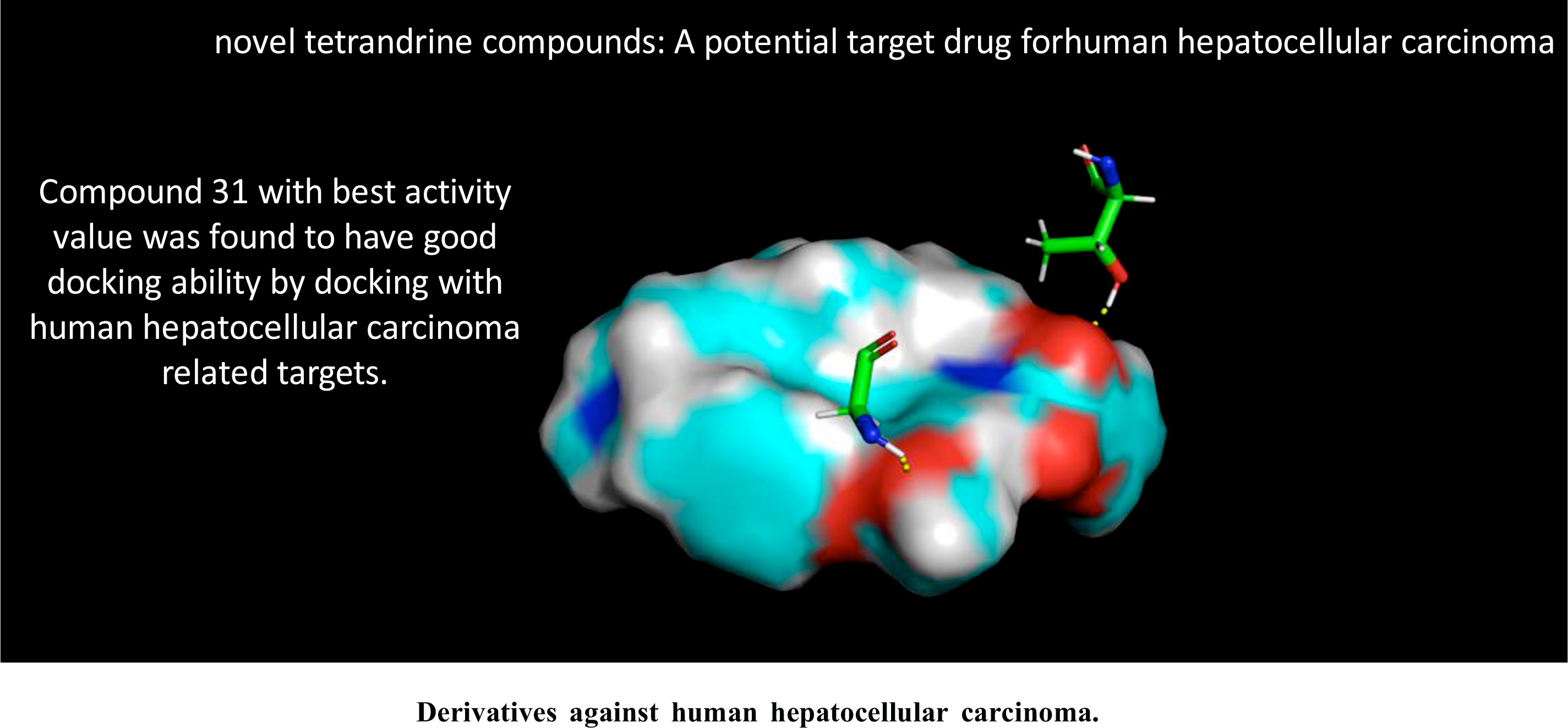
<P>Background: The new tetrandrine derivative is an anti-human liver cancer cell inhibitor which can be used to design and develop anti-human-liver-cancer drugs. <P> Objective: A quantitative structure-activity relationship (QSAR) model was established to predict the physical properties of new tetrandrine derivatives using their chemical structures. <P> Methods: The best descriptors were selected through CODESSA software to build a multiple linear regression model. Then, gene expression programming (GEP) was used to establish a nonlinear quantitative QSAR model with descriptors to predict the activity of a series of novel tetrandrine chemotherapy drugs. The best active compound 31 was subjected to molecular docking experiments through SYBYL software with a small fragment of the protein receptor (PDB ID:2J6M). <P> Results: Four descriptors were selected to build a multiple linear regression model with correlation coefficients R2, R2CV and S2 with the values of 0.8352, 0.7806 and 0.0119, respectively. The training and test sets with a correlation coefficient of 0.85 and 0.83 were obtained via an automatic problem-solving program (APS) using the four selected operators as parameters, with a mean error of 1.49 and 1.08. Compound 31 had a good docking ability with an overall score of 5.8892, a collision rate of -2.8004 and an extreme value of 0.9836. <P> Conclusion: The computer-constructed drug molecular model reveals the factors affecting the activity of human hepatocellular carcinoma cells, which provides directions and guidance for the development of highly effective anti-humanhepatocellular- carcinoma drugs in the future.</P>
Keywords:
Tetrandrine, structure-activity relationship, heuristic, gene expression, molecular docking, QSAR model.
Affiliation:
Graphical Abstract:
Read Full-Text article